Running Pipelines with the CLI¶
Capreolus provides a command line interface for running experiments using pipelines that are described by Task modules. To create a new pipeline, you’ll need to create a new Task before using the CLI.
Capreolus takes a functional approach to describing an experiment. An experiment is simply a pipeline plus a set of configuration options specifying both classes to use for the pipeline’s modules and configuration options associated with each module. These configuration options fully and deterministically describe the pipeline; the output should always be the same given the same configuration options (modulo any CUDA non-determinism). Capreolus takes advantage of this functional approach to cache intermediate outputs (given module dependencies).
Commands¶
The CLI takes a pipeline to run, such as rank.searcheval, and optionally a list of configuration options for the pipeline:
capreolus <pipeline> [with <configuration options>].
The first part of the pipeline corresponds to a Task (rank) and the second part corresponds to one of the Task’s commands (searcheval, which runs search followed by evaluate).
If no command is specified, a default chosen by the Task is run.
Configuration options are specified in key=value format.
All Tasks provide several commands to help understand their operation.
The print_config command displays the Task’s configuration, including any options specified on the command line.
The print_pipeline command displays the pipeline’s dependency graph, including current module choices.
Additionally, the modules Task provides list of all module types and classes that are currently registered. For example:
$ capreolus modules
module type=benchmark
name=antique
name=dummy
name=robust04.yang19
name=nf
...
module type=reranker
name=CDSSM
name=ConvKNRM
name=DRMM
name=DSSM
...
Example Pipelines¶
Note
Results and cached objects are stored in ~/.capreolus/results/ and ~/.capreolus/cache/ by default. Set the CAPREOLUS_RESULTS and CAPREOLUS_CACHE environment variables to change these locations. For example: export CAPREOLUS_CACHE=/data/capreolus/cache
RankTask¶
- Use
RankTaskwith the NFCorpusBenchmark. This will download the collection, create an index, and search for the NFCorpus topics. TheBenchmarkspecifies a dependency oncollection.name=nfand provides the corresponding topics and relevance judgments.
$ capreolus rank.searcheval with searcher.name=BM25 \
searcher.index.stemmer=porter benchmark.name=nf
- Use a similar pipeline, but with RM3 query expansion and a small grid search over expansion parameters. The evaluation command will report cross-validated results using the folds specified by the
Benchmark.
$ capreolus rank.searcheval with \
searcher.index.stemmer=porter benchmark.name=nf \
searcher.name=BM25RM3 searcher.fbDocs=5-10-15 searcher.fbTerms=5-25-50
RerankTask¶
- Use
RerankTaskto run the sameRankTaskpipeline optimized for recall@1000, and then train aRerankeroptimized for P@20 on the first fold provided by theBenchmark. We limit training to two iterations (niters) of sizeitersizeto keep the training process from taking too long.
$ capreolus rerank.traineval with \
rank.searcher.index.stemmer=porter benchmark.name=nf \
rank.searcher.name=BM25RM3 rank.searcher.fbDocs=5-10-15 rank.searcher.fbTerms=5-25-50 \
rank.optimize=recall_1000 reranker.name=KNRM reranker.trainer.niters=2 optimize=P_20
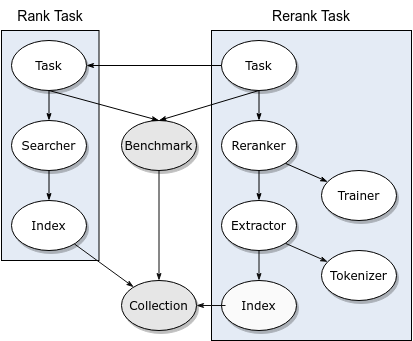
ReRerankTask¶
- The
ReRerankTaskdemonstrates pipeline flexibility by adding a second reranking step on top of the output fromRerankTask. Runcapreolus rererank.print_configto see the configuration options it expects. (Hint: it consists of aRankTasknamerankas before, followed by aRerankTasknamedrerank1, followed by anotherRerankTasknamedrerank2.)